Contents
Design of a selection for deoxycytidine deamination
PACE has enabled the rapid laboratory evolution of diverse protein functions, including protein–protein interactions35, tRNA synthetases36, DNA-binding proteins37,38,39, proteases40,41, polymerases42, metabolic enzymes43,44,45 and base editors7,12. During PACE, the evolving protein is encoded on the selection phage (SP), which infect Escherichia coli host cells46. The E. coli harbor a mutagenesis plasmid (MP) that constantly mutagenizes the phage genome as well as accessory plasmid(s) (AP) that establish a selection circuit that regulates the expression of gene III, which encodes pIII, a critical protein for phage replication. Because gIII has been removed from the SP genome, only phage that encode evolving variants with the desired activity trigger the production of pIII in E. coli and replicate, resulting in the propagation of active gene variants (Fig. 1b). Under constant mutagenesis and dilution, phage lacking the desired activity are rapidly diluted from the selection vessel (‘lagoon’), whereas phage that evolve beneficial mutations persist.
Previously, we developed a CBE-PACE selection12 in which a cytidine deaminase is encoded within the SP, and host E. coli cells contain (1) the MP, (2) an accessory plasmid that encodes SpCas9, (3) a self-inactivating T7 RNA polymerase (T7 RNAP) fused to a C-terminal degron and (4) gene III under T7 RNAP transcriptional control. Upon phage infection, the SP-encoded deaminase is joined to Cas9 by trans-intein splicing to reconstitute the base editor. To activate the selection circuit, the base editor must perform C•G-to-T•A editing to create a stop codon between T7 RNAP and the degron, yielding active T7 RNAP. Degron-free T7 RNAP then transcribes gIII, leading to phage propagation12.
To develop a PACE circuit to select for cytidine deamination by TadA, we modified our previous CBE selection circuit to accommodate an enzyme with high initial deoxyadenosine deamination activity (Fig. 1c). In the original circuit, TGG (Trp) is edited into a stop codon (TAG, TGA or TAA) through C-to-T conversion of CCA in the template strand. This strategy, however, places adenine, which is opposite thymine in all stop codons (TAG, TGA and TAA), at position 6 within the target protospacer. Given that position 6 is highly edited by ABE8e7, and that A-to-G editing of A6 precludes stop codon formation because CGG, CAG, CGA and CAA all encode amino acids, this original circuit would require high selectivity for deoxycytidine over deoxyadenosine deamination that is unlikely to be found among early-stage evolved ABE8e variants.
To address this problem, we developed a new selection circuit that instead edits the non-template strand (Fig. 1d). In the new circuit, C6A7A8 is edited to T6A7A8 to introduce a stop codon upon deoxycytidine deamination. Deoxyadenosine deamination does not prevent stop codon installation (TAA, TGA or TAG) in the new selection unless both A7 and A8 are converted to Gs (TGG = Trp), making this circuit tolerant to modest levels of deoxyadenosine deamination and, thus, more suitable for early-stage TadA8e evolution (Circuit 1). After initial evolution in the new circuit, we envisioned switching to the original template-strand circuit (Circuit 2) to take advantage of its inherent strong negative selection against deoxyadenosine deamination (Supplementary Fig. 1).
Deoxycytidine deaminase evolution
We initiated PANCE of TadA-8e using Circuit 1 (Fig. 1e). In PANCE, E. coli host cells containing the AP and MP are infected with phage containing the gene of interest and grown overnight, without continuous dilution. The next day, the supernatant containing the phage is diluted into a fresh host cell culture, and the process is repeated to enrich for phage harboring active cytidine deaminases. Compared to PACE, PANCE offers lower stringency and, thus, is helpful during early-phase evolution campaigns in which preserving genetically diverse variants with low initial activity can be critical7,41,43. After four rounds of PANCE with induced MP6 mutagenesis47, the phage began to propagate >100-fold overnight, suggesting improved activity for cytidine deamination. To increase the stringency of the selection, we increased the fold dilution between passages and decreased the strength of the promoter upstream of T7 RNAP (Supplementary Fig. 2). Next, we switched to Circuit 2 for additional passages of PANCE (Supplementary Fig. 2) to select against deoxyadenosine deamination while maintaining deoxycytidine deamination activity. To further increase selection stringency, we performed 159 hours of continuous evolution (PACE) on phage pools surviving PANCE using Circuit 2 (Supplementary Fig. 3). TadA-8e variants emerging from all phases of PANCE and PACE survived an average total dilution of ~10139-fold.
We isolated and sequenced individual phage surviving PANCE and PACE to identify TadA-8e mutations acquired during evolution (Fig. 2a and Supplementary Figs. 2 and 3). We observed a striking prevalence of mutations in residues 26–28 across all the sequenced phages, with R26G, E27K, E27A and V28G mutations highly represented across several separately evolved lagoons. Next, we assayed the evolved variants for base editing in E. coli. We sub-cloned five evolved TadA variants (TadA-CDa–e) from phage into the BE4max architecture48 (from N-terminus to C-terminus: TadA*–SpCas9–UGI–UGI) on a low-copy plasmid and designed a high-copy target plasmid containing sequences from the selection circuits on which the phage evolved. We co-transformed the base editor plasmid, which also encodes the guide RNA, and the target plasmid into E. coli cells, allowed editing after arabinose induction to occur overnight, and performed high-throughput sequencing of the target plasmid (Fig. 2b).
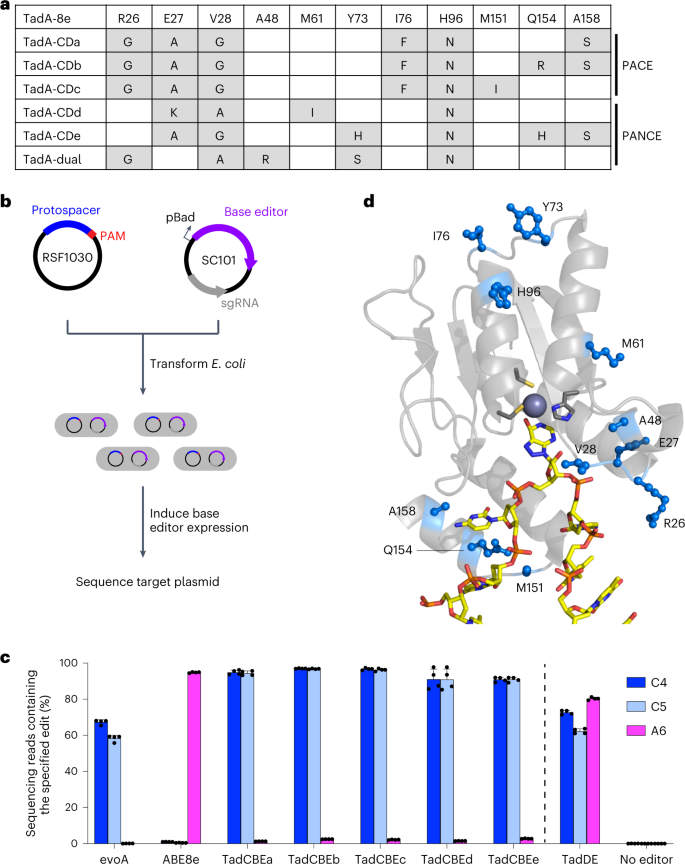
a, Summary of TadA-8e variants evolved and characterized in this work. The variants are representative of conserved mutations after nine passages of PANCE or after 159 hours of PACE. For a full list of mutations, see Supplementary Figs. 2 and 3. b, Method for assessing base editing of target plasmids in E. coli. Cells are co-transformed with a target plasmid (blue) and a base editor plasmid (purple). Base editor expression is induced with arabinose. After 16 hours, cells are harvested, and the target plasmid is analyzed by high-throughput sequencing. c, Base editing in E. coli of a protospacer matching the selection circuit target site. C•G-to-T•A edits are shown in blue. A•T-to-G•C edits are shown in magenta. Dots represent individual biological replicates, and bars represent mean ± s.d. from four independent biological replicates. d, Locations of evolved mutations in the cryo-EM structure of ABE8e (PDB: 6VPC)18.
Source data
The sequencing results revealed a striking shift in selectivity of the evolved TadA variants compared to the starting TadA-8e variant. Although base editors containing TadA-8e yielded 94% A•T-to-G•C editing at A6 and 1% C•G-to-T•A editing at C4 and C5 in the target plasmid, the evolved variants instead resulted in 90–97% editing of cytosines and 1–3% editing of adenine (Fig. 2c), representing a >3,000-fold change in cytosine versus adenine base editing. These results indicate that PANCE and PACE using selection Circuits 1 and 2 evolved TadA variants, hereafter referred to as TadA-cytidine deaminases (TadA-CDs), with strong cytidine deamination activity and high selectivity for cytosine over adenine base editing.
From a lagoon infected with TadA-8e A48R, containing a mutation that increases promiscuity in TadA-7.10 (ref. 32), we also identified a variant that performed both A•T-to-G•C (80%) and C•G-to-T•A (73%) editing in the E. coli editing assay (Fig. 2c). This variant thus serves as a TadA-based dual editor (TadDE). TadDE is smaller than previously reported dual editors that fuse both cytidine and adenosine deaminases to a Cas domain49,50,51,52,53 and may be especially useful for applications requiring broad mutagenesis54, such as genetic screens55,56.
To identify potential roles for the evolved mutations, we mapped them onto the cryogenic electron microscopy (cryo-EM) structure of ABE8e (Protein Data Bank (PDB): 6VPC)18. The highly conserved mutations are predicted to localize to a loop near the active site (Fig. 2d). This loop interacts with the backbone of the single-stranded DNA substrate near the target base and supports productive orientation of the base relative to the catalytic zinc ion. Other conserved mutations, including A158S and Q154R, also mapped to the interface of TadA and the single-stranded DNA substrate. A structural prediction of TadA-CDa using AlphaFold257,58 suggests that the mutations are not predicted to alter the structure of TadA compared to the cryo-EM structure of ABE8e (6VPC; Supplementary Fig. 4). Instead, the observed mutation of residues 26–28 from Arg-Glu-Val to smaller amino acids such as Gly-Ala-Gly during evolution may alleviate the steric clash that otherwise is predicted to block proper positioning of the pyrimidine C4 for nucleophilic attack and deamination (Supplementary Fig. 4). These observations collectively suggest that the evolved mutations may alter the conformation of the bound DNA substrate to enable efficient cytidine deamination and impede adenosine deamination.
We next performed mutagenesis and reversion analysis to interrogate the roles of the mutations found through evolution. In isolation, none of the mutations are sufficient to alter selectivity (Supplementary Fig. 5). However, the addition of just two mutations to the loop region (E27A V28G in TadCBEa–c,e and E27K V28A in TadCBEd) is sufficient to alter the selectivity of TadCBEs to modestly favor cytidine deamination, albeit with low editing efficiency (Supplementary Fig. 5). Additional mutations evolved during PANCE or PACE greatly increase activity and improve selectivity for C•G-to-T•A conversion. The reversion of mutations outside of the loop region generally decreases activity but not selectivity (Supplementary Fig. 6). This reversion analysis thus supports the importance of residues 26–28 in modulating the deamination selectivity of evolved TadA variants.
Characterization of TadA-CDs in mammalian cells
Encouraged by the characteristics of the TadA-CDs in bacteria, we evaluated the evolved TadCBEs in mammalian cells. We cloned five TadCBE variants (TadCBEa–e) into mammalian expression vectors regulated by a cytomegalovirus (CMV) promoter in the BE4max architecture48. These five TadCBE variants were assayed alongside three of the most widely used engineered and evolved CBEs: BE4max48, evoA12 and evoFERNY12. We co-transfected HEK293T cells with each base editor plasmid and a single guide RNA (sgRNA) plasmid, allowed editing to occur for 72 hours and then sequenced target sites from genomic DNA. Across nine different target sites tested in HEK293T cells, TadCBE variants generally yielded target C•G-to-T•A editing (averaging 51–60% peak editing for TadCBEa–e across all nine tested sites) that was similar to or higher than that observed from canonical BE4max, evoA and evoFERNY CBEs (averaging 47%, 55% and 41% peak editing, respectively, across all nine sites) (Fig. 3 and Supplementary Fig. 7). These results demonstrate that TadCBEs can perform highly efficient C•G-to-T•A editing in mammalian cells.
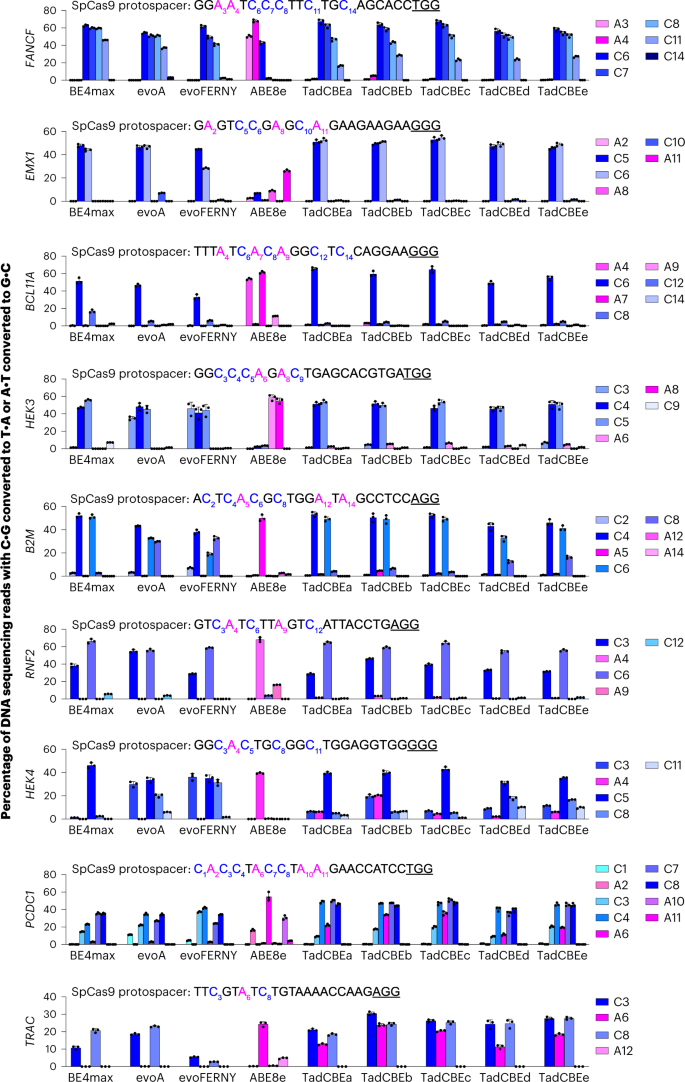
The specified base editors using SpCas9 nickase domains in the BE4max architecture or ABE8e with 2×UGI were transfected along with each of nine guide RNAs targeting the protospacers shown in each graph. Target cytosines are blue, target adenines are magenta, and PAM sequences are underlined. C•G-to-T•A base editing is shown in shades of blue. A•T-to-G•C base editing is shown in shades of magenta. Dots represent individual values, and bars represent mean ± s.d. of three independent biological replicates. HEK293T site 3 is abbreviated HEK3, and HEK293T site 4 is abbreviated HEK4.
Source data
Evolved TadCBE variants generally showed low residual A•T-to-G•C editing, averaging 1.5–4.5% editing for TadCBEa–e across adenosines in all nine tested sites and, thus, excellent selectivity for C•G-to-T•A editing over A•T-to-G•C editing (Fig. 3). By comparison, ABE8e in the same base editor architecture (with 2×UGI) averaged 31% A•T-to-G•C editing and 2.0% C•G-to-T•A editing across the nine sites. Ratios of desired C•G-to-T•A editing to residual A•T-to-G•C editing for seven of the nine tested sites was very high, averaging 21-fold to 42-fold for TadCBE variants a, c, d and e and 9.2-fold for TadCBEb (Fig. 3). Taken together, these observations suggest that residual A•T-to-G•C editing is generally low among evolved TadCBE variants, limited primarily to a small subset of target sites, protospacer positions and TadCBE variants. The introduction of V106W in the deaminase domain can further reduce residual A•T-to-G•C editing when necessary (vide infra).
On-target and off-target editing by TadCBEs
Highly active cytidine deaminases that natively modify DNA, such as APOBEC family enzymes, can deaminate transiently exposed single-stranded DNA beyond those in the R-loop defined by Cas9, leading to low-level but widespread Cas-independent modification of the genome13,14,15,19. Likewise, high-activity cytidine deaminases that can potently engage RNA can also mediate unguided off-target RNA deamination21. Cas-independent off-target DNA and RNA editing activity could limit the use of some CBEs in applications for which off-target editing must be minimized15. Cas-independent off-target DNA editing has been found to be undetected or much less frequent for several TadA*-based ABEs13, although overexpression of some ABEs can result in low-level RNA deamination6,7,34.
The TadA origin of TadCBEs offers several advantages for minimizing off-target editing, including the potential to include mutations that were found to reduce off-target DNA or RNA editing in previous TadA engineering efforts34,59,60. For ABEs, the addition of V106W to TadA-7.10, TadA-8e or TadA-8.17-m reduced Cas-independent off-target editing of DNA and RNA in all three cases while maintaining high levels of on-target activity6,7,34. We sought to test whether the V106W mutation when introduced into TadCBEs could reduce off-target DNA or RNA editing while maintaining on-target activity and selectivity. Because several evolved mutations in TadA-CDs are proximal to V106, it was not clear if the addition of V106W would disrupt desired TadA-CD properties (Supplementary Fig. 8).
We first evaluated the on-target activity of TadCBEs containing V106W. We constructed V106W variants of TadCBEa–e and evaluated editing efficiency at nine target sites in HEK293T cells. TadCBE variants a–e tolerated the addition of V106W and maintained high on-target cytidine deamination activity, averaging 56% peak C•G-to-T•A target editing efficiency across the nine tested target sites for TadCBEa–e V106W, nearly matching 57% average peak editing efficiency for TadCBEa–e (Fig. 4a and Supplementary Figs. 9–12). The TadCBEa–e V106W variants exhibited a slightly narrower editing window than TadCBEa–e while maintaining high peak editing efficiency (Supplementary Fig. 12). Encouragingly, cytosine versus adenine base editing selectivity was improved 3.1-fold on average for TadCBE V106W variants compared to the corresponding TadCBE variants across these nine sites (Supplementary Fig. 12). TadCBE-V106W variants, thus, can retain efficient cytosine base editing with improved selectivity for deoxycytidine over deoxyadenosine deamination and refined editing windows.
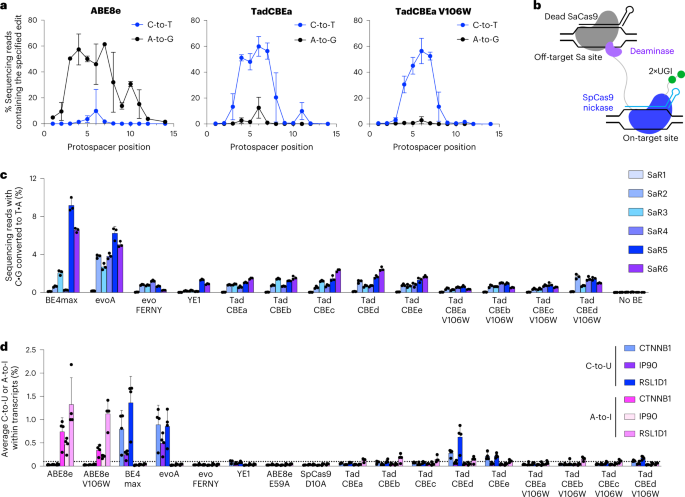
a, Base editing activity window for ABE8e with 2×UGI, TadCBEa and TadCBEa V106W across nine different target genomic sites in HEK293T. Dots represent average editing across all sites containing the specified base at the indicated position within the protospacer. Individual data points used for this analysis are in Fig. 3 and Supplementary Figs. 9 and 11. b, Method for measuring Cas-independent off-target DNA editing with the orthogonal R-loop assay. c, Average Cas-independent off-target editing across all cytosines within six orthogonal R-loops (SaR1–SaR6) generated by dead S. aureus Cas9. d, Off-target RNA editing. RNA was harvested from HEK293T cells 48 hours after transfection with the indicated base editor. After cDNA synthesis, CTNNB1, IP90 and RSL1D1 were amplified and analyzed by high-throughput sequencing. For c and d, dots represent individual biological replicates, and bars represent mean ± s.d. of three (c) or four (d) independent biological replicates.
Source data
Next, we evaluated Cas-independent DNA editing by TadCBEs and TadCBE-V106W variants using the previously established orthogonal R-loop assay15,19 (Fig. 4b). This assay measures the propensity of a base editor to modify single-stranded DNA in an off-target R-loop generated by an orthogonal, catalytically inactive Staphylococcus aureus Cas9 (SaCas9). By sequencing genomic DNA across six unrelated off-target SaCas9 R-loops, we determined that TadCBEs, on average, have 3.7-fold lower Cas-independent off-target C•G-to-T•A editing (0.84%–1.2%) compared to BE4max (3.6%) and evoA (3.8%) (Fig. 4c and Supplementary Figs. 13–16). The average off-target activity of evoFERNY (0.58%) and YE1 (0.53%) were also low. The addition of V106W further reduced Cas-independent off-target editing of TadCBEs by an average factor of 1.9 (to 0.38%, 0.62% 0.48%, 1.1% and 0.11% for V106W TadCBE variants a–e, respectively). Consistent with the selectivity of TadCBEs for deoxycytidine deamination, we did not detect appreciable off-target A•T-to-G•C editing by any TadCBEs (Supplementary Fig. 17). These findings indicate that evolved TadCBEs have inherently low Cas-independent off-target DNA editing that can be further suppressed by adding V106W while retaining high on-target C•G-to-T•A editing and low residual A•T-to-G•C editing.
We also evaluated off-target RNA editing by TadCBEs (Fig. 4d and Supplementary Figs. 18 and 19). After transfection of HEK293T cells by TadCBEa–e, BE4max, evoA, evoFERNY, ABE8e or ABE8e-V106W, RNA was extracted from cells. After complementary DNA (cDNA) synthesis, three target transcripts (CTNNB1, IP90 and RSL1D1) previously used to measure off-target RNA editing due to their abundance or sequence similarity to the native TadA tRNAArg2 substrate2,15,19,34 were amplified by RT–PCR and analyzed for C-to-U or A-to-I editing by high-throughput sequencing. Although BE4max and evoA edited, on average, ~0.7% of the analyzed cytosines in these transcripts, evoFERNY, YE1, TadCBEa, TadCBEb and TadCBEc all edited ≤0.1% of the cytosines (our limit of detection) (Fig. 4d and Supplementary Fig. 18). TadCBEd and TadCBEe edited, on average, 0.3% and 0.2% of cytosines across the three transcripts, respectively. The addition of V106W reduced average off-target RNA editing down to ≤0.13% in both cases (Fig. 4d and Supplementary Fig. 18).
Taken together, these data suggest that TadCBEs offer much lower frequencies of Cas-independent off-target DNA and RNA editing compared to BE4max and evoA. Off-target editing by TadCBEs is substantially less frequent than that of any other CBE of similar on-target activity and size. When further reduction of off-target editing is essential, the addition of V106W minimizes off-target DNA and RNA editing, focuses the editing window to ~4–5 base pairs and minimizes residual deoxyadenosine deamination, with only a small reduction in maximal on-target activity.
Finally, Cas-dependent off-target editing occurs when base editors engage a non-target site that resembles the target site through imperfect Cas9 binding61. We analyzed Cas-dependent off-target activity in HEK293T cells at 22 known off-target sites for SpCas9 base editors and sgRNAs targeting HEK293T site 3 (hereafter referred to as HEK3), HEK293T site 4 (hereafter referred to as HEK4), EMX1 and BCL11A (Supplementary Figs. 20–25). Across multiple validated off-target sites, we observed that Cas-dependent off-target editing by TadCBEs was generally similar to the low level observed for BE4max and evoA variants (Supplementary Figs. 20–25). The Cas-dependent off-target activity of YE1 and evoFERNY was still lower, consistent with the lower on-target activity of these variants (Supplementary Figs. 20–25).
Collectively, these findings suggest that TadCBEs offer lower Cas-independent off-target DNA and RNA editing compared to canonical CBEs and low levels of Cas-dependent off-target DNA editing consistent with those observed for currently used CBEs of similar on-target editing efficiencies. The use of high-fidelity Cas proteins that engage fewer off-target loci is known to reduce Cas-dependent off-target DNA base editing62, and their use in TadCBEs may offer the same benefits.
Characterization of TadCBEs on 10,638 target sites
TadCBE activity can vary substantially by target site (Fig. 3). To comprehensively characterize the activity of TadCBEs across a wide range of sites in mammalian cells, we performed high-throughput analysis of base editing outcomes for TadCBE variants using our previously reported ‘comprehensive context library’ of 10,638 paired sgRNA and target sites integrated into an mESC line (Supplementary Fig. 26)11. These libraries include target sites with all possible 6-mers surrounding a substrate A or C nucleotide at protospacer position 6 and all possible 5-mers across positions −1 to 13 (counting the position immediately upstream of the protospacer as position 0) with minimal sequence bias11. Base editing conditions were optimized to allow differences between base editors to be detected. We maintained an average cell coverage of ≥300× per library member throughout the course of the experiment and an average sequencing depth of ≥2,800× per target, which enabled us to detect editing outcomes with high sensitivity. We collected two biological replicates per base editor for TadCBEa–e, V106W variants of TadCBEa–d, TadDE, and BE4max as a reference11, and validated that the library assay data have strong consistency between biological replicates (Supplementary Fig. 27).
We used the resulting library data to quantify editing activity and C•G-to-T•A selectivity for each TadCBE (Fig. 5a). Across the 10,638 integrated target sites, all TadCBE and TadCBE-V106W variants edited with greater average efficiency (28–31% of reads on average with any C•G-to-T•A editing) than BE4max (21%) (Fig. 5a)11. We next characterized the editing windows, which we defined as positions within the protospacer that averaged ≥30% of the peak average editing efficiency (Fig. 5b and Supplementary Fig. 28). TadCBE editing is generally centered around protospacer position 6. The most active variant, TadCBEd, has a similar editing window (protospacer positions 3–9) to that of BE4max (positions 3–9), whereas the remaining TadCBEs and V106W-TadCBEs have slightly narrower windows (positions 3–8; Fig. 5b and Supplementary Fig. 28).
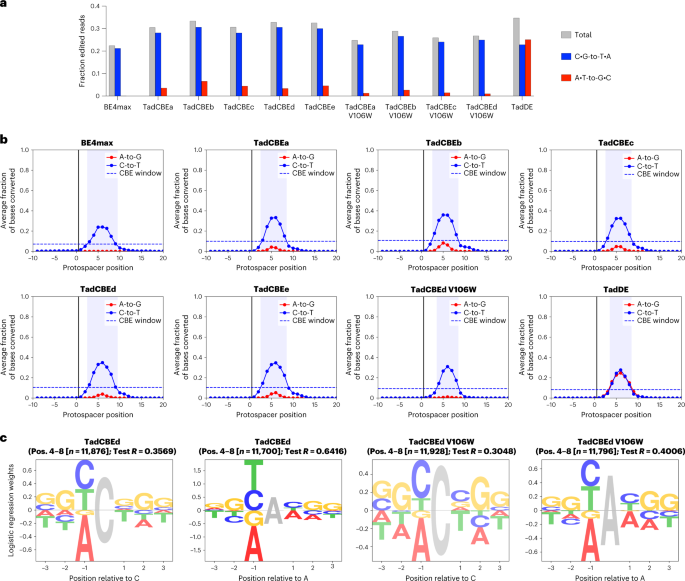
a, Overall efficiency and selectivity of base editors analyzed through editing of the library. Data show the average fraction of edited sequencing reads across all library members between protospacer positions −9 to 20, where positions 21–23 are the PAM. b, BE4max, TadCBEa–e, TadCBEd V106W and TadDE editing profiles across 10,638 genomically integrated target sites. The editing window is defined as the protospacer positions for which average editing efficiency is ≥30% of the average peak editing efficiency. Window plots for all variants tested in the library experiment can be found in Supplementary Fig. 28. c, Sequence motifs of TadCBEd and TadCBEd V106W for cytosine and adenine base editing outcomes determined by performing regression on editing efficiencies. Opacity of sequence motifs is proportional to the test R on a held-out set of sequences. Complete sequence motif plots for all variants are in Supplementary Fig. 30.
Source data
TadCBE selectivity for cytosine editing over adenine editing varied by base editor. Among the canonical TadCBEs (without V106W), TadCBEd showed the highest C•G-to-T•A selectivity, with a geometric mean of the ratio of C•G-to-T•A versus A•T-to-G•C editing at each position in its editing window of 26.8 (Methods and Supplementary Table 1). Notably, the addition of V106W substantially improved C•G-to-T•A selectivity for all TadCBE variants (TadCBEd V106W selectivity = 47.8) while minimally affecting base editing activity at the maximally edited position (Fig. 5a,b and Supplementary Fig. 29). For example, the addition of V106W to TadCBEd reduced peak editing among the library targets from 35% to 31%.
Consistent with the discrete target site examples shown above, C•G-to-T•A selectivity of TadCBEs varied by target site across the comprehensive context library. Adenine base editing was observed in 3.4–6.6% of reads (on average) for TadCBEs and 1.0–2.7% of reads for V106W-TadCBEs across all target sites in the comprehensive context library (Fig. 5a). We generated sequence motifs by performing regression on the editing efficiencies to determine the sequence characteristics that affect cytosine and adenine deamination (Methods, Fig. 5c and Supplementary Fig. 30). TadCBEs have similar sequence context preferences to their ancestor, ABE7.10, favoring editing of cytosine and adenine bases preceded by 5′ Y (Y = T/C) while disfavoring 5′ A (ref. 11). When performing bystander adenine base editing, TadCBEs retain the sequence context preference of ABE7.10 (favoring 5′ YAY and disfavoring 5′ AAA). However, TadCBEs instead slightly disfavor 5′ ACT. The difference in 3′ preference may be due to differences in substrate positioning required to achieve altered selectivity, because interactions with adjacent bases could alter placement of the target cytidine in the active site (Supplementary Fig. 4).
TadDE performs very similar levels of adenine and cytosine base editing (ABE:CBE ratio = 1.1) and has similar sequence context dependence to TadCBEs (Fig. 5a,b, Supplementary Fig. 30 and Supplementary Table 1). TadDE is highly efficient, editing 35% of the reads on average in the library experiment (Fig. 5a). The probability of observing A•T-to-G•C editing given that C•G-to-T•A editing is observed is 0.62 for TadDE compared to 0.04 for TadCBEd-V106W, our most selective TadCBE variant (Supplementary Table 1). The high activity, promiscuity and small size of TadDE makes it a promising tool for concurrent A•T-to-G•C and C•G-to-T•A editing.
Collectively, these data show that TadCBEs have greater cytosine deamination activity than conventional narrow-window CBEs. Furthermore, the introduction of V106W in the deaminase domain reduces residual A•T-to-G•C editing activity while minimally impacting C•G-to-T•A editing for all TadCBEs in this experiment. Overall, TadCBEd enables the greatest cytosine deamination activity with high C•G-to-T•A selectivity, which is further improved by the addition of V106W.
TadCBE compatibility with Cas9 orthologs
The use of Cas9 orthologs with diverse PAM requirements expands the targetable sequence space of base editors. To test if TadCBEs are compatible with Cas9 homologs beyond Streptococcus pyogenes Cas9, we constructed TadCBE variants with PACE-evolved variants of Nme2Cas9 from Neisseria meningitidis that broaden the scope of accessible PAMs beyond the canonical NGG PAM of SpCas9 (ref. 63). We recently evolved Nme2Cas9 variants that access a wide range of single-pyrimidine PAM sites as nucleases or as base editors64. We generated fusions of TadA-CDs with an eNme2-C variant nickase (PAM = N4CN) and two UGI domains, co-transfected the resulting eNme2-C-TadCBEs with a guide RNA plasmid and examined base editing at six genomic loci in HEK293T cells. Across all tested sites, the peak editing efficiency of TadCBEs was similar to that of BE4max, evoFERNY and evoA (Supplementary Figs. 31 and 32). Although C•G-to-T•A editing exceeded 50% at some sites, residual A•T-to-G•C editing never exceeded 5.3% at any of the six eNme2 target sites tested. TadCBEs thus exhibited robust activity and selectivity with eNme2-CCas9 variants.
We next tested TadCBEs with SaCas9 in the BE4max architecture65. SaCas9 (1,053 amino acids) is smaller than SpCas9 (1,368 amino acids) and recognizes a different PAM sequence (PAM = NNGRRT). We found that TadCBEs using SaCas9 have robust C•G-to-T•A editing across nine sites (4.1–44%) with less than 5.5% A•T-to-G•C editing at any site (Supplementary Figs. 33 and 34). These observations suggest potential compatibility with other Cas proteins that, together with SpCas9, eNme2-C Cas9 and SaCas9, may offer access to a variety of PAM sequences for versatile targeting of TadCBEs. We additionally found that TadDE performed both A•T-to-G•C and C•G-to-T•A editing with SpCas9, eNme2-C Cas9 and SaCas9 in mammalian cells at sites where TadCBEs were selective, suggesting broad Cas9 compatibility of the dual editor as well (Supplementary Figs. 31–36).
TadCBEs exhibit a narrower editing window than BE4max, evoA and evoFERNY CBEs while maintaining similar or higher maximal editing efficiencies (Supplementary Fig. 31). For example, BE4max and evoA edited Neisseria meningitidis site 50 (hereafter referred to as Nme50) at protospacer positions 3–18 with 4.2–47% efficiency, whereas TadCBEa, TadCBEb and TadCBEc modify only the narrower position 3–8 window with 5–48% efficiency (Supplementary Fig. 31). The narrower base editing activity window of TadCBEs could arise from a less processive deaminase, because the processive APOBEC family deaminases can catalyze multiple hydrolytic deamination reactions per DNA-binding event66. Although a wide editing window can be useful for some applications, such as targeted gene disruption or base editing screens, the narrower window of TadCBEs should benefit precision editing applications in which modification of only one target base is desirable, particularly when using Cas9 domains that support a wider base editing window63,67. Taken together, the small size of TadCBEs, their compatibility with eNmeCas9 and SaCas9, their more focused editing windows and their high editing efficiencies and selectivities for cytosine over adenine base editing demonstrate their suitability for a variety of precision cytosine base editing applications.
Multiplexed base editing in primary human T cells
We evaluated whether TadCBEs can perform multiplexed editing of target loci in T cells in support of therapeutic applications. Multiplexed base editing in T cells can modify or disrupt multiple genes with minimized risks of chromosomal abnormalities and cell state perturbations that arise from multiple double-stranded breaks68,69,70,71,72. To determine whether TadCBEs can perform multiplexed editing in primary human T cells, we targeted the CXCR4 and CCR5 loci for simultaneous base editing to install premature stop codons in both HIV co-receptors (Fig. 6a,b)73. We performed in vitro transcription (IVT) of TadCBE variants a, b, c, d and e. We then electroporated the TadCBE mRNA along with guide RNAs targeting CXCR4 and CCR5 (Fig. 6a,b)73 into primary human T cells and analyzed editing efficiencies at both target sites.

mRNA encoding the indicated base editor or GFP as a negative control was electroporated into human T cells (n = 4 donors) along with two synthetic guide RNAs targeting CXCR4 (a) or CCR5 (b) at the specified protospacers. Target cytosines are blue, target adenines are magenta, and PAM sequences are underlined. After 3 days, genomic DNA was harvested from T cell lysates and analyzed by high-throughput sequencing. The gray boxes indicate the desired location of stop codon installation in CXCR4 and CCR5. The targeted cytidine to yield TAG (CXCR4) and TAA (CCR5) stop codons upon cytosine base editing is underlined. c, mRNA encoding the indicated base editor or GFP as a negative control was electroporated into HSPCs along with a synthetic guide RNA targeting the BCL11A enhancer. After 3 days, genomic DNA was harvested from cell lysates and analyzed by high-throughput sequencing. C•G-to-T•A base editing is shown in shades of blue, and A•T-to G•C-base editing is shown in shades of magenta. Dots represent individual biological replicates, and bars represent mean ± s.d. from n = 4 donors (a and b) or n = 3 donors (c).
Source data
TadCBEs performed efficient (averaging 70%) and selective editing of the target cytosines (C7 in CXCR4 and C9 in CCR5), resulting in premature stop codon installation in each gene (Fig. 6a,b). Editing efficiencies of TadCBEs were similar to those of BE4max (67%) and evoA (76%) (Fig. 6a,b). Observed indel frequencies of all the tested base editors was comparably low (typically ≤0.68%; Supplementary Fig. 37). Consistent with data in HEK293T cells (Supplementary Fig. 12), TadCBEs exhibited a more precise editing window with fewer bystander edits at CXCR4 and CCR5 in primary human T cells. Because TadCBEs maintain high editing efficiencies and product purities but offer substantially lower Cas-independent off-target DNA and RNA editing than APOBEC and evoA (Fig. 4c,d and Supplementary Figs. 13–17), TadCBEs provide a promising alternative for multiplexed cytosine base editing of T cells.
We also compared T cell editing by TadCBEs to that of evoFERNY and YE1, which offer similarly low off-target editing as TadCBEs (Figs. 6a,b and 4c,d and Supplementary Figs. 13–17). TadCBEs supported substantially higher editing efficiencies in T cells than evoFERNY and YE1. At CXCR4, target C•G-to-T•A editing efficiency by TadCBEs averaged 1.5-fold to 1.7-fold that of evoFERNY and YE1, whereas, at CCR5, average TadCBE editing efficiencies were 4.9-fold to 11-fold higher on average. We analyzed three known Cas-dependent off-target sites for the CCR5 guide RNA and one known off-target for CXCR4 and found that Cas-dependent off-target editing was lower for TadCBEa–e, evoFERNY and YE1 (≤0.12%) than for BE4max (0.1–0.58%) and evoA (0.1–1.0%) (Supplementary Fig. 38). Next, we tested V106W variants of TadCBEa–e in T cells. Relative to their TadCBE counterparts, the V106W variants displayed 1.3-fold to 1.9-fold lower average activity at C7 of CXCR4 and 1.4-fold to 3.3-fold lower average activity at C9 of CCR5, with a proportional drop in C•G-to-G•C editing (Supplementary Figs. 39–41). These data are consistent with the narrower editing window of V106W variants and suggest that the more transient mRNA delivery of TadCBEs may reveal a greater range of editing activity compared to plasmid transfections of HEK293T cells. Overall, these findings demonstrate that TadCBEs offer a favorable combination of on-target and off-target editing features compared to currently used CBEs when base editing primary human T cells at target sites of therapeutic relevance.
Editing in human HSPCs
Finally, we evaluated the editing efficiency of TadCBEs in human HSPCs. We electroporated TadCBEa–e mRNA along with a synthetic guide targeting the enhancer of BCL11A into primary human CD34+ cells. Mutations at the enhancer can decrease the expression of BCL11A, leading to induction of fetal hemoglobin expression as a potential treatment for sickle cell disease74,75. For comparison, we electroporated mRNA encoding BE4max, evoA, evoFERNY, YE1 or GFP (as a negative control) in parallel. evoFERNY and YE1 yielded only 2.7% and 2.0% average editing, respectively, whereas BE4max and evoA averaged 7.0% and 7.4% editing efficiencies, respectively (Fig. 6c). All five of the tested TadCBEs supported 2–3-fold-higher editing efficiencies than BE4max or evoA, averaging 14–23% (Fig. 6c). All of the tested CBEs yielded low levels of indels (≤1.1%; Supplementary Fig. 42a) and Cas-dependent off-target editing (≤0.87%; Supplementary Fig. 42b). These results demonstrate that the editing efficiencies of TadCBEs can exceed that of the most commonly used CBEs for some therapeutically relevant sites and cell types.




